Connecting Rice Germplasm to Plant Breeding: Backcrossing for Allele Mining and Recurrent Selection for Allele Pyramiding Through Molecular Marker Technology
Received: 31-Mar-2014 / Accepted Date: 02-Apr-2014 / Published Date: 04-Apr-2014 DOI: 10.4172/2329-8863.1000e114
Green Revolution
‘Green revolution’ in 1960s and successful utility of heterosis in 1970s have resulted in two successive leaps in rice productivity [1,2]. Although extensive breeding efforts were made for high yield in past decades, the yield potential of modern rice varieties has remained stagnant for many years. However, the world’s rice production need to double again by the year 2030 to keep up with the demands of a growing population [3] and much of this increase will mainly depend on improved rice cultivars. Therefore, further improving yield potential has been the top priority in almost all rice breeding programs worldwide.
QTL Mapping and Allele Mining by Advanced Backcross Method
The rich genetic diversity in the worldwide rice germplasm collections contains genes capable of improving almost all traits of modern varieties. Favorable genes affecting yield and other agronomic traits can be identified by molecular genetic maps [4]. Recently, a wide range of segregating populations derived from bi-parental crosses, including recombinant inbred lines (RILs), doubled haploid lines (DH), F2 and its derived populations, and BC or testcross populations, have been used for mapping QTL [5]. Huge numbers of genes/QTL have been identified and mapped on the 12 rice chromosomes (http://www.grammene.org/). However, two factors may be contributing to the less-than-expected impact of marker-based QTL analysis on the development of varieties with enhanced quantitative traits:
(1) Allele diversity can’t be dissected and analyzed due to segregation of only two alleles at each locus in the bi-parental derived populations, resulting in failure to mine favorable or novel alleles from germplasm. The phenotypic effects of different alleles in rice germplam at those identified QTL and their values in rice breeding remain largely unknown, making it difficult to use them in breeding.
(2) Separation of QTL mapping from breeding practice. Because QTL expression largely depends on genetic background and environment, QTL detected in the mapping populations can’t be directly applied in the breeding population.
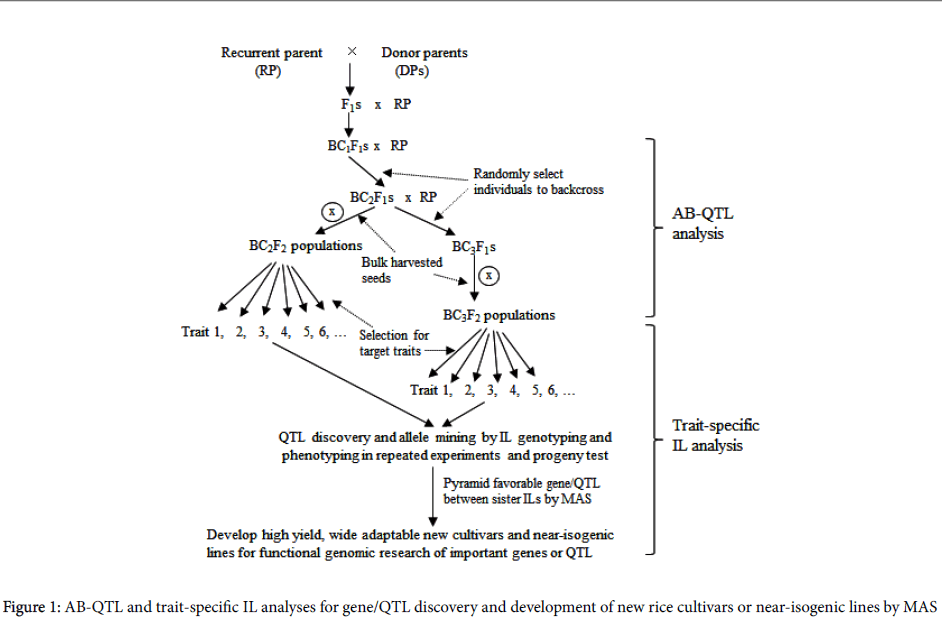
To integrate QTL mapping with improvement of rice variety, Tanksley and Nelson [4] proposed the method of advanced backcross QTL (AB-QTL) analysis as a way to control background genetic variation in QTL mapping (Figure 1). By simultaneously identifying and transferring favorable QTL alleles from unadapted to cultivated germplasm, AB-QTL analysis has since become a popular way of genetic dissection of quantitative trait variation in many crops such as rice [6,7] maize [8], wheat [9] and barley [10].
The advantages of advanced populations over balanced populations (e.g., F2, BC1, RILs) are: (1) Accuracy in measurement of yield and other traits in similar background with the elite variety, thus ensuring QTL mapping results more meaningful. (2) Less epistasis detected due to the overall lower frequency of donor alleles in advanced generations, making a better prediction of the identified QTL when it is transferred into the cultivated background. (3) High efficient integration of QTL discovery with variety improvement in the same population. However, AB-QTL analysis also faces the challenge of incapability of mining allele diversity and high cost of genotyping probably resulting from the large BC mapping population derived from bi-parents.
Subsequently, a strategy of genome-wide trait-specific introgression lines (ILs) was proposed that had got widely used in molecular dissection of complex phenotypes and pyramiding of favorable alleles for enhanced quantitative traits in rice [10-13], which is consisted of three steps (Figure 1):
(1) Development of advanced backcross populations. Series of introgressed bulk populations can be developed in elite variety’s backgrounds using diverse germplasm accessions as donor parents by backcross breeding procedure.
(2) Trait screening and progeny test. BC bulk populations will be screened and progeny-tested for target traits such as high yield, resistances or tolerances to different abiotic and biotic stresses, resulting in multiple sets of trait-specific introgression lines (ILs), which much outperform the recurrent parents for the target traits.
(3) Gene/QTL detection and allelic mining. Gene/QTL detection can be conducted by tracking and characterizing genome-wide responses of donor segments to selection. Deviation (excess or deficiency) of donor allele frequency at single loci in ILs from the expected level estimated in the specific random BC populations can be detected by X2 tests [12]. Favorable alleles for the target traits will be identified by comparisons of gene effects of different alleles from diverse donors in the same elite background. Based on phenotypic performance and distribution of favorable alleles of ILs, it is easy to pyramid different favorable alleles in the same elite background by marker-assisted selection (MAS).
Although analogous to the AB-QTL analysis approach, the trait-specific ILs mapping strategy was more time-saving and cost-effective than the conventional QTL mapping approach in at least three aspects:
(1) Selecting extreme phenotypes visually was much easier than phenotyping the whole population practiced in typical QTL mapping experiments, thus minimizing errors in QTL mapping resulting from phenotyping fluctuation.
(2) A small number of ILs with extreme phenotypes selected from each BC population greatly reduces the genotyping and phenotyping cost.
(3) Trait-specific ILs are actually target-trait improved materials in the elite background with many trait-enhanced alleles from diverse donors, providing an ideal platform of materials for rice molecular breeding.
Favorable allele pyramiding by recurrent selection
Chronic crossing with high pressure of selection within populations derived from elite varieties has been a mainstream method of modern breeding in crops. The pedigree method usually involves a limited number of elite progenitors-frequently related to each other – that are crossed. The long-term risk with pedigree selection is a reduced genetic base and small genetic gains.
Owing to unfavorable linkage drag from exotic germplasm, many main-effect or major QTL identified from wild rice haven’t been successfully utilized in rice breeding programs. This is a reason why inbred variety hasn’t made breakthrough in yield over few past decades. Developing populations with broad genetic bases and using improvement methods that permit the continuous accumulation of favorable alleles can overcome these disadvantages.
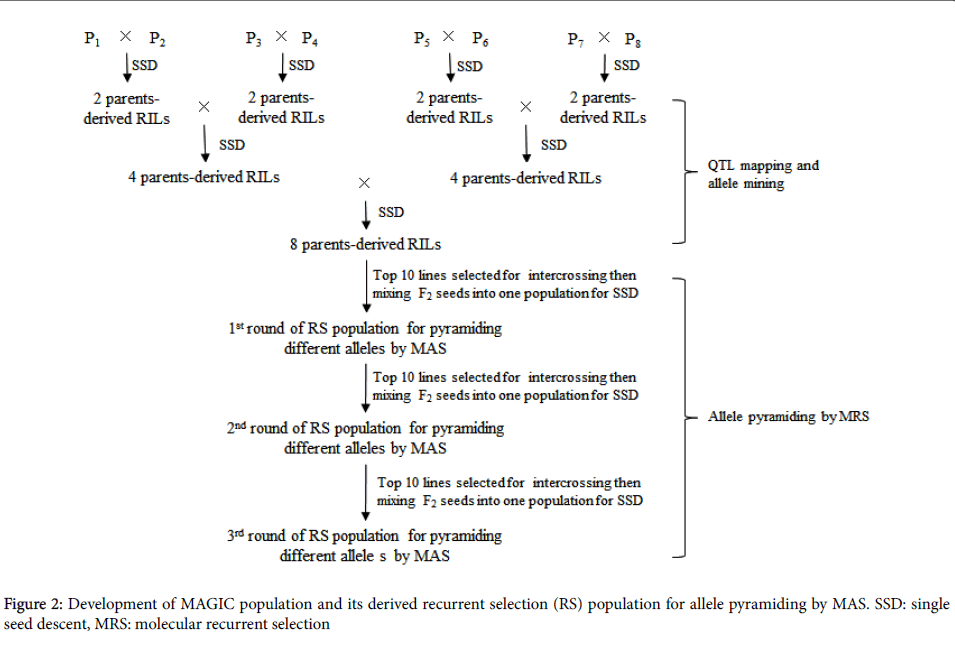
Recurrent selection (RS) can fit population improvement by high efficient pyramiding of different favorable alleles from diverse donors. Recently, multi-parents advanced generation inter-cross (MAGIC) population has been introduced to plant community [14]. MAGIC population is of characteristics of diversities both in alleles and phenotypes which are from multiple parents. In addition, relatively large recombination events within the population ensure resolution of QTL mapping.
Based on QTL mapping information and phenotypic performance, ideal individuals will be selected by pyramiding of multiple favorable alleles in the consecutive recurrent selection populations derived from intercrossing among the top plants in each round of RS (Figure 2). MAGIC populations have been developed for the purpose of integration of QTL mapping with breeding in rice [15] and wheat [16] crops.
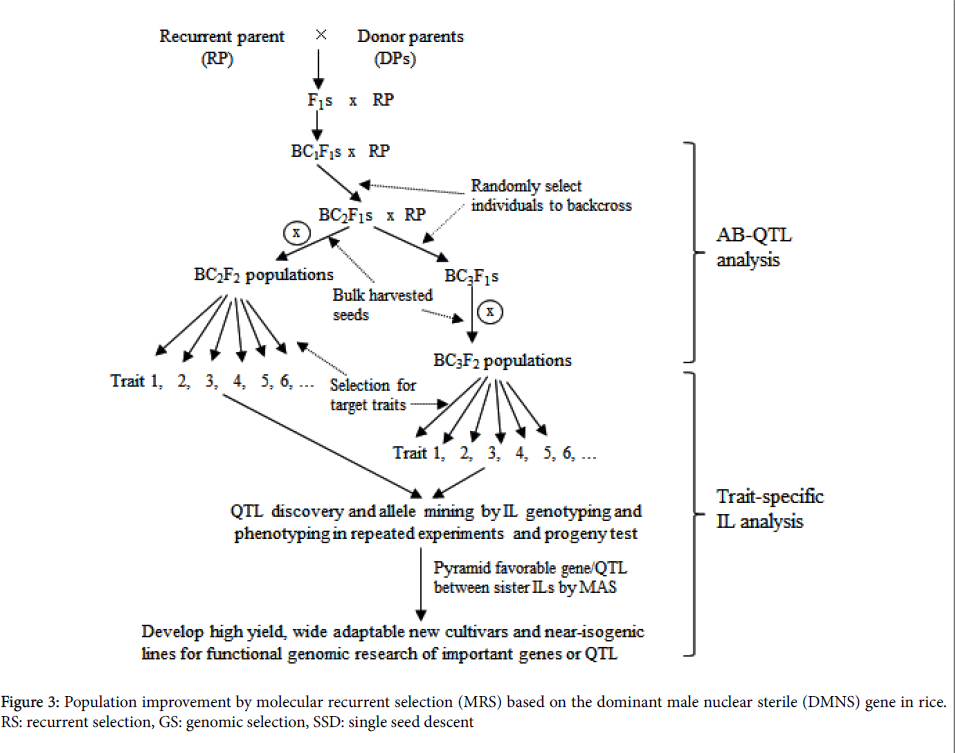
Discovery and introgression of a dominant male nuclear sterile (DMNS) gene [17] into elite backgrounds make easy recombination of different alleles, producing half sterile individuals and another half fertile individuals in each round RS population. Human-aided outcrossing between DMNS sterile plants and fertile plants greatly facilitates recombination of alleles at all loci among individuals in the progeny of the RS population.
The strategy of molecular marker combined with RS, referred to as molecular recurrent selection (MRS), probably provides a very promising way to pyramid different favorable alleles (Figure 3). It should allow more effective to identify recombinant of favorable alleles at all target loci and more effective breakdown of genetic drags in the MRS population through DMNS facilitated random mating. Furthermore, genomic selection (GS) can be entirely integrated with RS.
Bulk-harvested seeds from all fertile plants in the RS population after the third round of recombination could be used for a training population after genetic uniformity is achieved by rapid generation advancement (RGA) facilitation. Based on phenotypic data in the given environment and high throughput SNP data of the training population, QTL affecting the target traits will be identified and genetic prediction model will be set up for each target trait (Figure 3). In the subsequent RS populations, the prediction model can be used to estimate genomic breeding values (GEBV) of individuals for selection based on marker data, thus making more accuracy in selection owing to avoiding phenotyping of the target traits.
In practice, both approaches separately involving promising trait-specific ILs in an elite variety background and diverse elite varieties can be used as base parents to cross with DMNS. As compared with the latter, the former has an advantage of synchronous heading date within a RS population which ensures random outcrossing of individuals with the DMNS plants but a relative disadvantage of lesser diversity in its subsequent RS populations.
In brief, rapid development of high throughput phenotyping and genotyping technologies has been facilitating identification of QTL affecting important agronomic traits in crops [18]. Backcrossing for allele mining and RS for allele pyramiding by MAS are becoming a high efficient way to integrate QTL mapping with breeding practice. It will greatly promote crop molecular breeding from concept to practice.
References
- Pingali PL (2012) Green revolution: impacts, limits, and the path ahead. Proc Natl Acad Sci USA 109:12302–12308.
- Yuan LP (2001) Breeding of super hybrid rice. In: S Peng, B Hardy (eds), Rice Research for Food Security and Poverty Alleviation. Edited by International Rice Research Institute 143–149.
- Foley JA, Ramankutty N, Brauman KA, Cassidy ES, Gerber JS, et al. (2011) Solutions for a cultivated planet. Nature 478:337-342.
- Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 277: 1063-1066.
- PatersonAH (1996) Mapping genes responsible for differences in phenotype.In: AH Paterson (ed), Genome Mapping in Plants,RG Landes Company, Austin41–54.
- Moncada P, Martínez CP, Borrero J, Chatel M, GauchJrH, et al. (2001) Quantitative trait loci for yield and yield components in an Oryza sativa × O. rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet 102:41-52.
- Thomson MJ, Tai TH, McClung AM, Lai X-H, Hinga ME, et al. (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479-493.
- Ho JC, McCouch SR, Smith ME (2002) Improvement of hybrid yield by advanced backcross QTL analysis in elite maize. Theor Appl Genet 105:440-448.
- Huang XQ, Cöster H, Ganal MW, Röder MS (2003) Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.). Theor Appl Genet 106:1379-1389.
- Pillen K, Zacharias A, Léon J (2004) Comparative AB-QTL analysis in barley using a single exotic donor of Hordeum vulgare ssp. spontaneum. Theor Appl Genet 108:1591-1601.
- Ali AJ, Xu JL, Ismail AM, Fu BY, Vijaykumar CHM, et al.(2006) Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program. Field Crops Research 97: 66-76.
- Li ZK, Fu BY, Gao YM, Xu JL, Ali J, et al.(2005) Genome-wide introgression lines and a forward genetics strategy for functional genomic research of complex phenotypes in rice. Plant Molecular Biology 59:33-52.
- Li ZK, ZhangF (2013) Rice breeding in the post-genomics era: from concept to practice. Current Opinion in Plant Biology 16: 261-269.
- Cavanagh C, Morell M, Mackay I, Powell W (2008) From mutations to MAGIC, resources for gene discovery, validation and delivery in crop plants. Curr Opin Plant Biol 11: 215-221.
- Bandillo N, Raghavan N, Muyco PA,Sevilla MAL, Lobina IT, et al. (2013) Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6: 11-26.
- Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ,et al. (2012)Amultiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnology Journal 10: 826-839.
- Huang XB, Tian ZH, Deng ZQ, Zheng JT, Lin CB, et al. (2008) Preliminary identification of a novel Sanmingdominant male sterile gene in rice (Oryza sativaL.). Acta Agronomica Sinica 34: 1865-1868.
- Cabrera-BosquetL, CrossaJ, von ZitzewitzJ, SerretMD,ArausJL (2012) High-throughput phenotyping and genomic selection: The frontiers of crop breeding converge. Journal of Integrative Plant Biology 54:312- 320.
Citation: Jian-Long Xu and Jauhar Ali (2014) Connecting Rice Germplasm to Plant Breeding: Backcrossing for Allele Mining and Recurrent Selection for Allele Pyramiding Through Molecular Marker Technology. Adv Crop Sci Tech 2:e114. Doi: 10.4172/2329-8863.1000e114
Copyright: © 2014 Jian-Long X, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 16020
- [From(publication date): 8-2014 - Apr 27, 2024]
- Breakdown by view type
- HTML page views: 11259
- PDF downloads: 4761